Most research disciplines, regardless of whether in STEM or social sciences, consist of a common approach — taking a specific methodology or approach, applying some computation to analyze whatever data are collected to gain insights into a new topic and draw conclusions. How these approaches and analyses are recorded and made available is critical to assess, reproduce, and fully understand the research, any resulting data and insights.
When research is publicly funded, there is a solid argument for what’s produced to be openly available and the need for open research suddenly gains clarity and focus during a global public health crisis. Health professionals around the world need, and absolutely should have, access to any new scientific diagnostic approaches, as quickly as possible. Emerging or trusted computational tools for modeling disease epidemiology, or approaches for machine learning prognosis using clinical characteristics, should be made available for immediate reuse by others.
A global scientific collaboration has been lauded as scientists come together to perform research during the pandemic. The New York Times piece, “Covid-19 Changed How the World Does Science, Together” noted:
“While political leaders have locked their borders, scientists have been shattering theirs, creating a global collaboration unlike any in history. Never before, researchers say, have so many experts in so many countries focused simultaneously on a single topic and with such urgency. Nearly all other research has ground to a halt.”
Many funders and health organizations are demanding that research approaches and results be made open. Preprints have offered one solution, and their value during this challenging time has been evident in the huge volume of COVID-19 related content appearing online. For example, this collection of COVID-19 SARS-CoV-2 preprints on medRxiv and bioRxiv has more than 1900 manuscripts.
Now, protocols.io and Code Ocean are working to ensure that those research approaches remain open. These open access online tools are ideal repositories for all protocol and methodological approaches as well as computational pipelines and code. Online collaborative research tools are helpful to researchers who are restricted in how they can work and collaborate. For those at the frontline conducting scientific research, these tools serve as an ideal way to share their insights and approaches. Here’s how protocols.io and Code Ocean are supporting the research community during this unprecedented time:
protocols.io
A Dedicated Open Methods Community in protocols.io
When methods and protocols relating to the SARS-CoV2 virus and COVID-19 started to appear on protocols.io earlier this year, the protocols.io team quickly realized the value of creating the “Coronavirus Method Development Community”, a shared workspace for new protocols relevant to the virus. An incredibly fast-growing community on protocols.io, this Open Community already has more than 300 members and is growing daily.
On the Coronavirus Method Development Community, many members are active, adding comments to already posted protocols or starting their own discussion threads. The protocols are many and varied, from COVID-19 clinical diagnostic PCR approaches, viral plaque assays, methods to culture SARS-CoV-2, and Coronavirus sequencing protocols (some of which were adapted as new ‘forked’ versions of Ebola sequencing protocols that were already available on protocols.io). Other protocols include mask building ideas, how to 3D print personal protective equipment and the World Health Organization’s recommended hand-rub formulations.
When asked about using protocols.io during the current crisis, Public Health Virologist Ian MacKay, a member of the protocols.io open community noted the following:
“protocols.io makes it possible to rapidly put our tested methods onto an open-access platform. It permitted ongoing curation of the protocol, and it provided us a platform for further discussion with the ability to really drill down into the nitty-gritty about the protocol with other users, if needed. We used protocols.io for our SARS-CoV-2 PCR methods as soon as we could. We wanted those who might not have access to expertise in assay design, optimization and validation so they could benefit from our work. protocols.io provides a global collegiality that is in dire need at times like these.”

protocols.io Editorial Support
Alongside, and in support of, this motivated community, protocols.io has been working behind the scenes to grow and mobilize their team of editors to support researchers offering to import and check coronavirus-related protocols into the open community, then assigning the protocol back to the appropriate contact person.
Code Ocean
Since the outbreak of COVID-19, the team at Code Ocean has announced unlimited cloud compute and storage for COVID-19-related projects. Code Ocean’s goal has always been to help accelerate the pace of research by providing researchers with a better way to develop, collaborate and share code and data. Science builds on transparency, replication and collaboration. Code Ocean is currently supporting researchers working on COVID-19 in the following two areas:
Rapid dissemination of interactive and open research results
Code Ocean provides in-browser access to compute capsules, permanent records of pre-built computational environments containing all software, code, data, and results for a project. Compute capsules allow researchers to instantly inspect and interact with published work, as well as dynamically update new code or datasets to explore new possibilities based on existing research.
One recent compute capsule accompanying a medRxiv preprint from the Global Policy Lab at The University of California, Berkeley is an example of the rapid and open research response to COVID-19. Hsiang et al. evaluated the effect of large-scale anti-contagion policies on the COVID-19 pandemic. The analysis includes 49 packages and code in Python, R and Stata, and public epidemiological datasets from 6 countries, all of which is open access and fully reproducible. The study finds:
“We estimate that, to date, current policies have already prevented or delayed on the order of eighty-million infections. These findings may help inform whether or when ongoing policies should be lifted or intensified, and they can support decision-making in the over 150 countries where COVID-19 has been detected but not yet achieved high infection rates.”
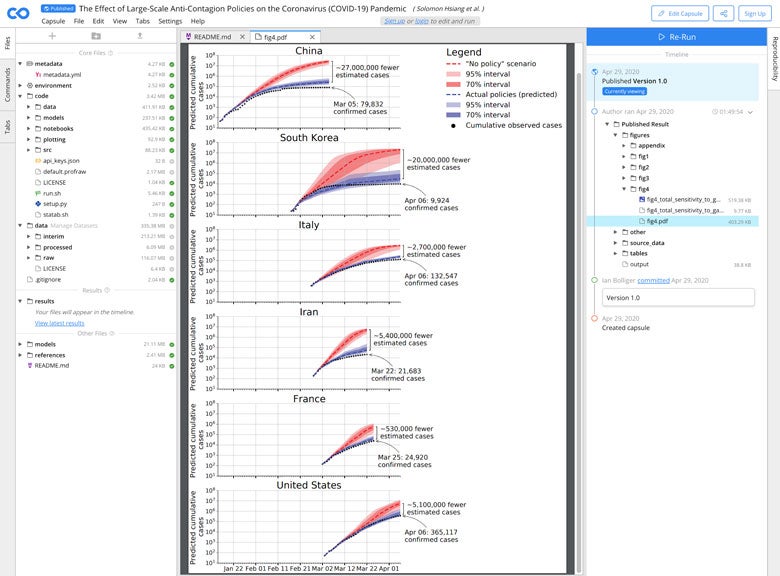
As the authors state in the capsule:
“The easiest way to interact with our code and data is via this Code Ocean capsule, because all of the relevant setup described below has been done for you.”
When a research project is published in a compute capsule, researchers interested in the work can directly re-run the analysis interactively in a built-in computational workbench, including a command line interface, JupyterLab/Notebook, RStudio, and Shiny on cloud instances with a few clicks on Code Ocean. Alternatively, researchers can also export the entire capsule and rebuild the project (using a Docker image) on their own devices without a Code Ocean account.
All capsules preserve the data used in the analysis even if this data is removed from the original public location in the future. Some capsules, including the example from Hsiang et al. above, also allow users to dynamically update the analysis with new data as they are being released.
Whether in-browser or rebuilt locally, interactive compute capsules can lower the barrier to examine and extend new research findings, potentially shortening decision-making cycles and helping with time-sensitive policies.
Convenient access to preconfigured cloud compute environments
In countries, campuses or labs experiencing disruptions or without access to research compute infrastructure, Code Ocean provides a web-based solution that can be accessed with only a laptop and the internet. Algorithms, simulations, and analyses in different programming languages can be easily shared and multiple collaborators added, enabling teams to continue research activities at a distance.
One difficulty with internal and external collaborations is onboarding collaborators. With Code Ocean, teams do not have to spend time setting up computational environments. Instead, new team members with different levels of technical experience can get started in the same computational environment right away.
For a complex disease like COVID-19, it takes time to gather the various pieces of the puzzle from researchers around the globe. Unfortunately, we’re in a race against time to better understand the pathology, the genetics that underpin the disease, and how best to control and treat, or vaccinate against COVID-19. Researchers need to avail themselves of tools that already exist to support and accelerate their ability to make progress in these vital efforts.
The combination of Code Ocean and protocols.io, along with other open tools, could significantly shorten the cycle to disseminate each advance and make the resulting efforts and insights reproducible. These tools reduce the iterations needed to confirm and build on results, whether formally published or preprinted, and they reduce the effort required to validate existing or emerging methods. Making the most of the combined power offered by research infrastructure tools that support and capture the development and dissemination of computational and/or experimental research processes, and facilitate research being built upon, will accelerate the overall pace of scientific discovery.
